AI tool set to transform characterization and treatment of cancers
A multinational team of researchers, co-led by Prof. Smita Krishnaswamy of Yale, has developed and tested a new AI tool to better characterize the diversity of individual cells within tumors, opening doors for more targeted therapies for patients.
"Our study is the first time that single cell data have been able to simplify this continuum of cell states into a handful of meaningful archetypes through which diversity can be analysed to find meaningful associations with spatial tumour growth and metabolomic signatures,” says Krishnaswamy, associate professor of computer science and genetics. "This could be a game changer."
Findings on the development and use of the AI tool, called AAnet, have today been published in Cancer Discovery.
Read more • Approximately 5 minutes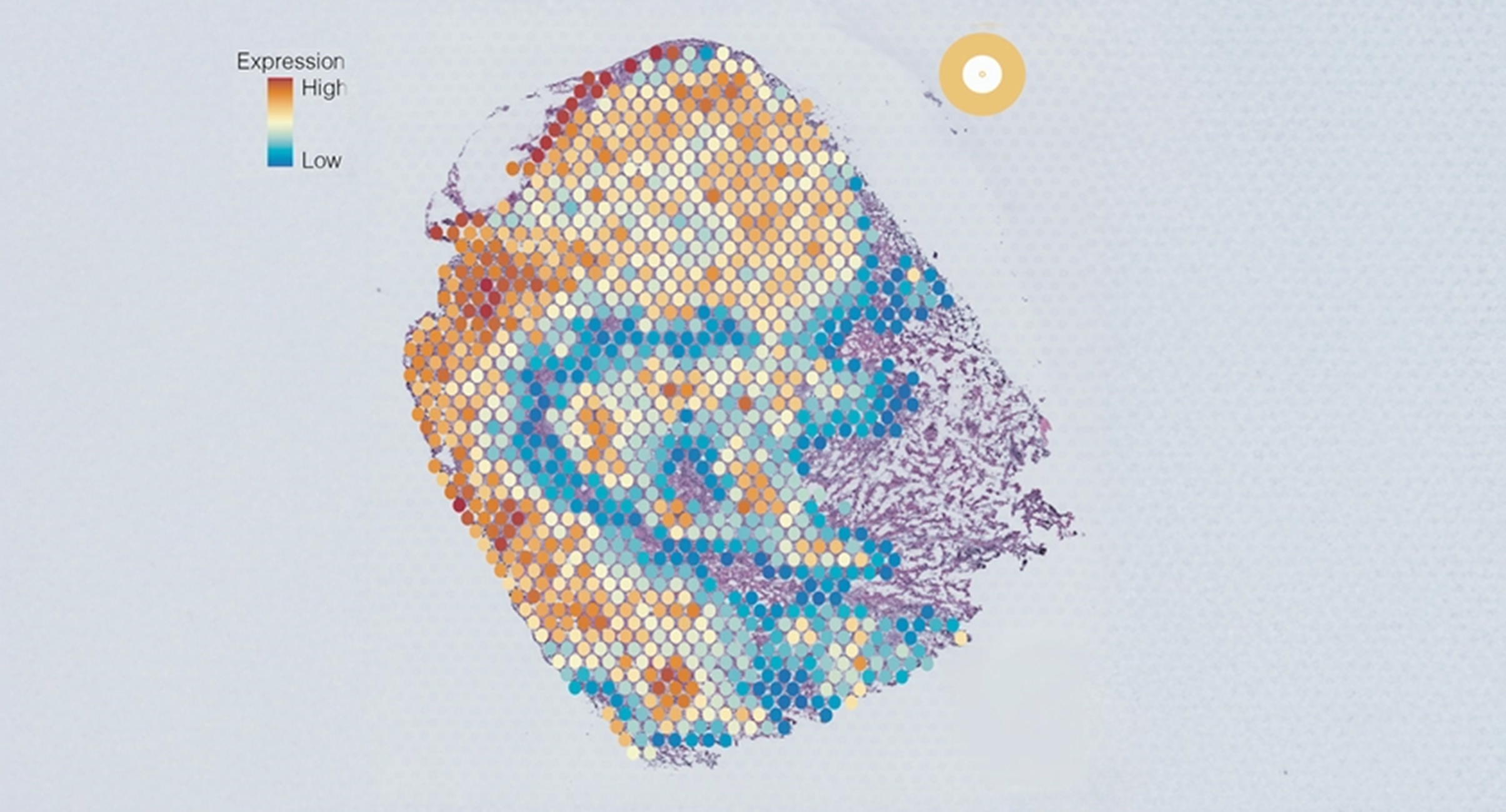
Tumor cross section showing five cell groups, each colored differently based on gene expression.
Not all tumor cells the same
Tumors aren't made of just one cell type – they're a mix of different cells that grow and respond to treatment in different ways. This diversity, or heterogeneity, makes cancer harder to treat and can in turn lead to worse outcomes, especially in triple negative breast cancer.
“Heterogeneity is a problem because currently we treat tumors as if they are made up of the same cell. This means we give one therapy that kills most cells in the tumor by targeting a particular mechanism. But not all cancer cells may share that mechanism. As a result, while the patient may have an initial response, the remaining cells can grow and the cancer may come back,” says associate professor Christine Chaffer, co-senior author of the study and co-director of the Cancer Plasticity and Dormancy Program at the Garvan Institute of Medical Research.
But while heterogeneity is a problem, researchers don’t know enough to characterize it: “So far researchers haven’t been able to clearly explain how adjacent cells in a tumor differ from each other, and how to classify those differences into meaningful ways to better treat tumors. But this is exactly what we need to know so we can kill all cells within that tumor with the right therapies,” Chaffer adds.
A new tool characterizes five new cancer cell groups

“With this data we have been finding out that not only is each patient’s cancer different, but each cancer cell behaves differently from another.
Smita Krishnaswamy
Associate professor of computer science and genetics
To solve this problem, the team developed and trained a powerful new AI tool called AAnet that can detect patterns in data of individual cells within tumors. They used the AI tool to uncover patterns in the level of gene expression of individual cells within tumors to derive new groups. The team focused on human models of triple-negative breast cancer and human samples of ER positive, HER2 positive and triple-negative breast cancer. Through this, they identified five different cancer cell groups within a tumor, with distinct gene expression profiles that indicated vast differences in cell behavior.
“By using our AI tool, we were consistently able to discover five new groups of cell types within single tumors called ‘archetypes’. Each group exhibited different biological pathways and propensities for growth, metastasis and markers of poor prognosis. Our next steps are to see how these groups may change over time, for example before and after chemotherapy,” says Chaffer.
Having access to this level of detail is a major breakthrough for cancer research.
“Thanks to technology advances, the last 20 years have seen an explosion of data at the single cell level," Krishnaswamy said. "With this data we have been finding out that not only is each patient’s cancer different, but each cancer cell behaves differently from another."
New classification to drive better, targeted treatments
The researchers say the use of AAnet to characterize the different groups of cells in a tumour according to their biology opens doors for a paradigm shift in how we treat cancer.
“Currently the choice of cancer treatment for a patient is largely based on the organ that the cancer came from such as breast, lung or prostate and any molecular markers it may exhibit. But this assumes that all cells in that cancer are the same. Instead, now we have a tool to characterize the heterogeneity of a patient’s tumour and really understand what each group of cells is doing at a biological level. With AAnet, we now hope to improve the rational design of combination therapies that we know will target each of those different groups through their biological pathways. This has the potential to vastly improve outcomes for that patient,” says Chaffer.
On the application of AAnet, co-senior author of the study and Chief Scientific Officer of Garvan Professor Sarah Kummerfeld states: “We envision a future where doctors combine this AI analysis with traditional cancer diagnoses to develop more personalised treatments that target all cell types within a person’s unique tumour. These results represent a true melding of cutting-edge technology and biology that can improve patient care. Our study focused on breast cancer, but it could be applied to other cancers and illnesses such as autoimmune disorders. The technology is already there.”
This research was supported through the following sources.
In Australia: The NELUNE Foundation, Tour de Cure, Estee Lauder, The Kinghorn Foundation, The Paramor Family Foundation, University of New South Wales Scientia Research Fellowship, Ramaciotti Biomedical Research Award , ARC Development Project grant and NHMRC Ideas Grants and Investigator Grant.
In the US: Gruber Foundation Science Fellowship and the Eric and Wendy Schmidt Center at the Broad Institute of MIT and Harvard , National Science Foundation , Yale Cancer Center Pilot grant and Sloan Fellowship.
More Details
Published Date
Jun 24, 2025


